Reconstructed State Space
Below is an example of how to create a recurrence network based on a reconstructed state space. We’ll examine the variables restless and procrast.
#----------------------
# Adjacency matrix
#----------------------
library(casnet)
p1 <- est_parameters(y = out.cart$restless)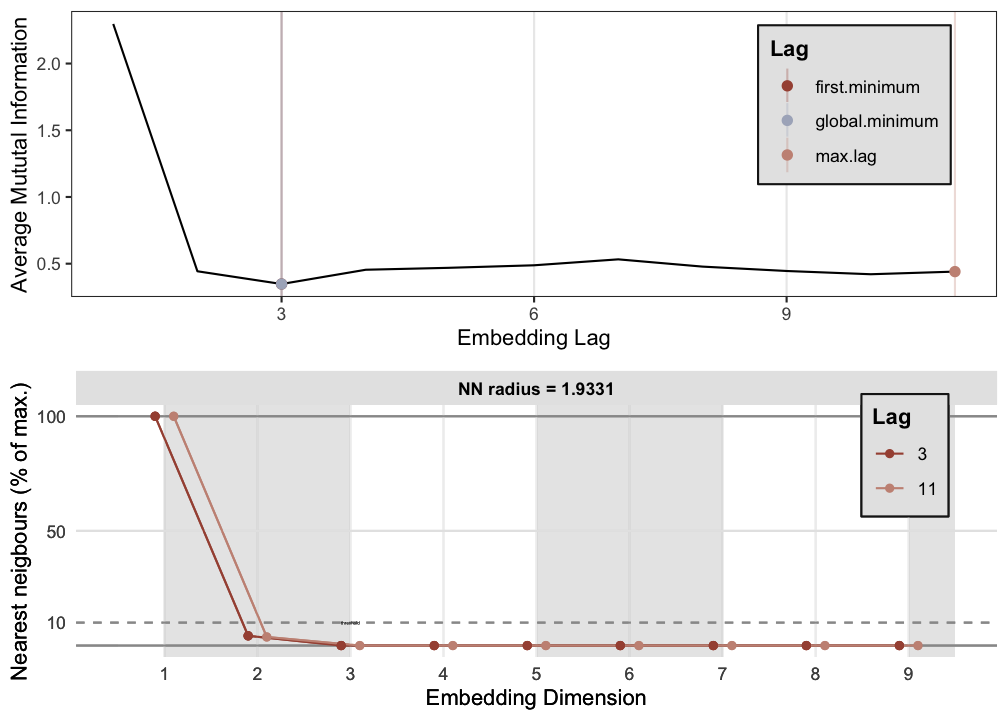
p2 <- est_parameters(y = out.cart$procrast)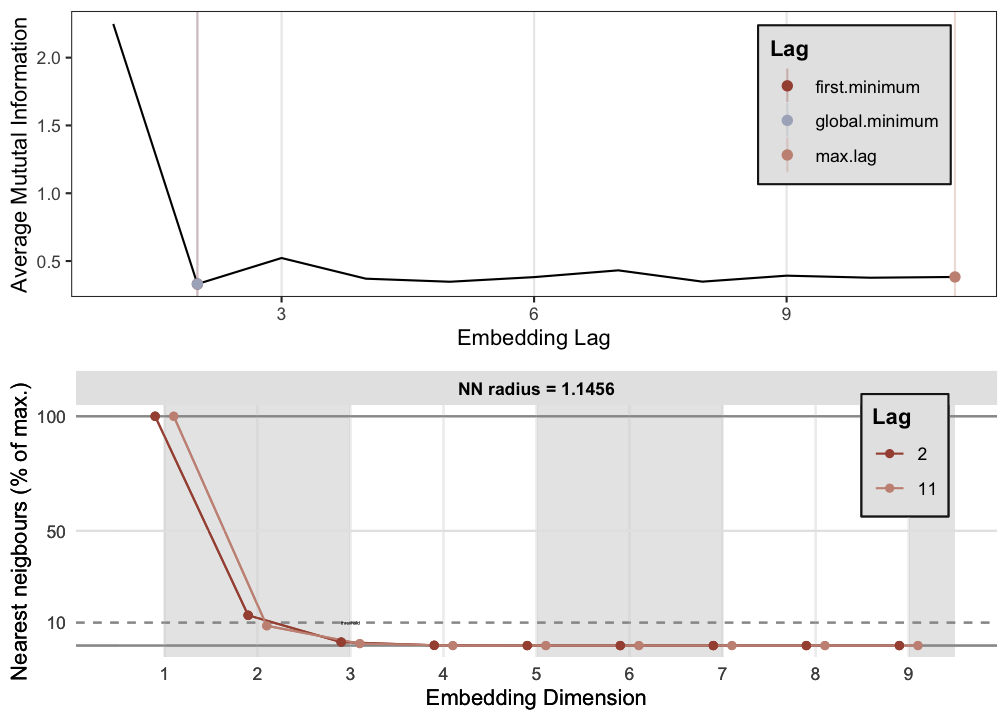
# By passing emRad = NA, a radius will be calculated based on 5% recurrence
RN1 <- casnet::rn(y1 = out.cart$restless, emDim = p1$optimDim, emLag = p1$optimLag, emRad = NA)
casnet::rn_plot(RN1)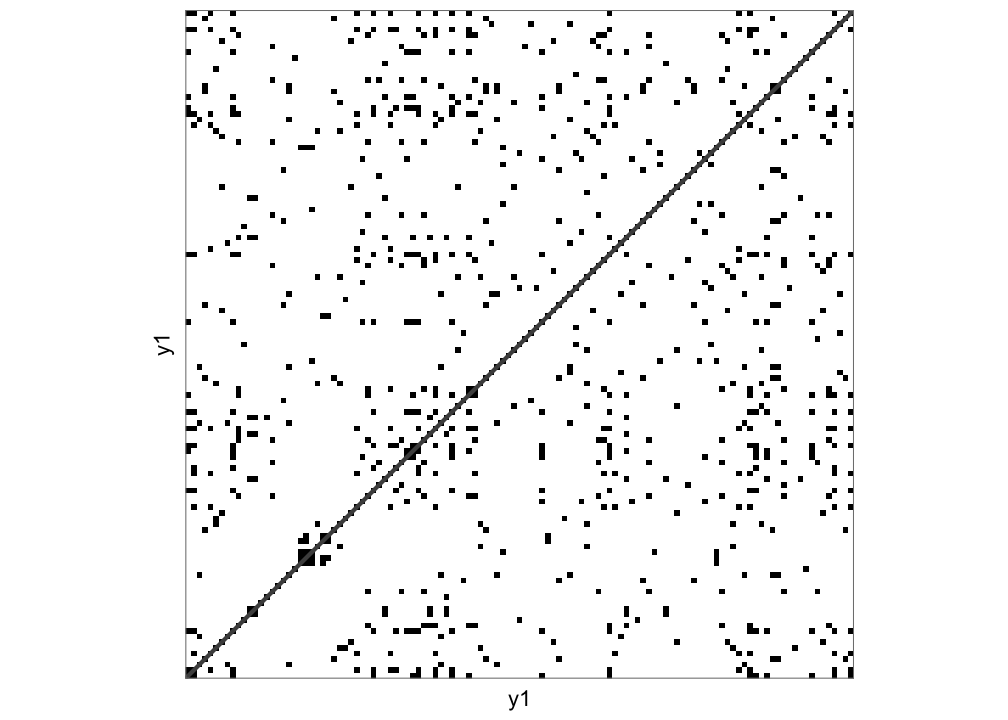
# Get RQA measures
rqa1 <- casnet::rp_measures(RN1, silent = FALSE)>
> ~~~o~~o~~casnet~~o~~o~~~
> Global Measures
> Global Max.points N.points RR Singular Divergence Repetitiveness
> 1 Matrix 14161 887 0.0626 688 0.0084 1.65
>
>
> Line-based Measures
> Lines N.lines N.points Measure Rate Mean Max. ENT ENT_rel CoV
> 1 Diagonal 41 199 DET 0.224 4.85 119 0.115 0.0240 3.76
> 2 Vertical 76 164 V LAM 0.185 2.16 3 0.436 0.0913 0.17
> 3 Horizontal 76 164 H LAM 0.185 2.16 3 0.436 0.0913 0.17
> 4 V+H Total 152 328 V+H LAM 0.185 2.16 3 0.436 0.0913 0.17
>
> ~~~o~~o~~casnet~~o~~o~~~# Create RN graph
g1 <- igraph::graph_from_adjacency_matrix(RN1, mode="undirected", diag = FALSE)
igraph::V(g1)$size <- igraph::degree(g1)
g1r <- casnet::make_spiral_graph(g1,
markEpochsBy = weeknum[1:vcount(g1)],
markTimeBy = TRUE)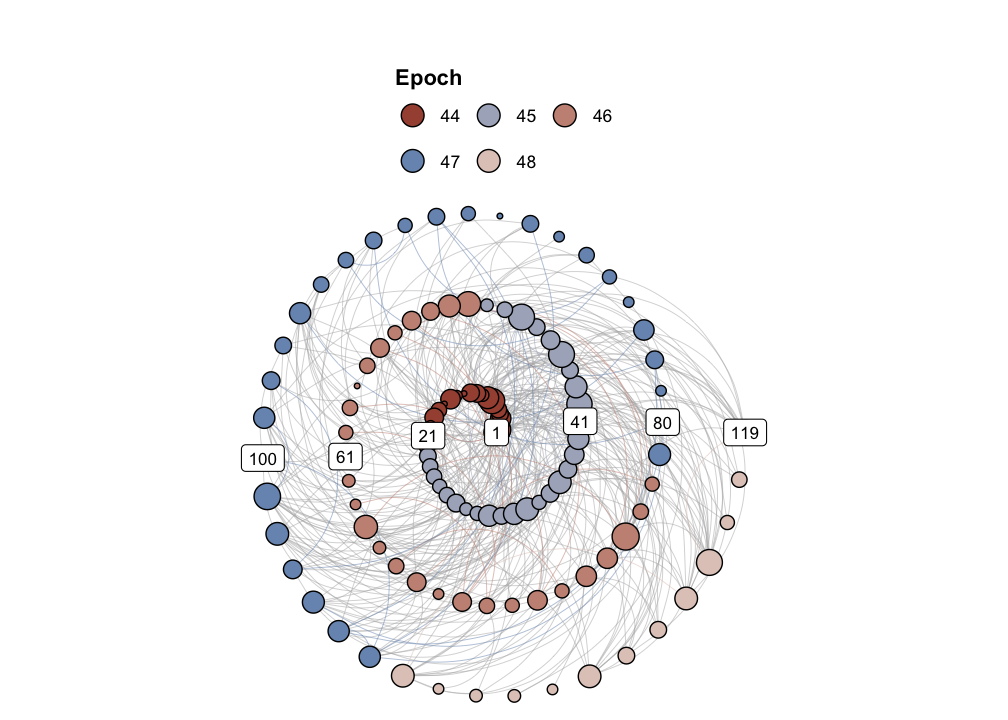
# Get RN measures
rn1 <- rn_measures(g1, silent = FALSE)>
> ~~~o~~o~~casnet~~o~~o~~~
>
> Global Network Measures
>
> EdgeDensity MeanStrengthDensity GlobalClustering NetworkTransitivity
> 1 0.05469306 NA 0.5443074 0.6294523
> AveragePathLength GlobalEfficiency
> 1 Inf 7.585679
>
> ~~~o~~o~~casnet~~o~~o~~~# Should be the same
rqa1$RR==rn1$graph_measures$EdgeDensity> [1] FALSETS 2
#----------------------
# Adjacency matrix TS_2
#----------------------
plot(ts(series$TS_2))
# Because these are generated signals, look for a drop in FNN below 1%.
p2 <- est_parameters(y = series$TS_2, nnThres = 1)
RN2 <- rn(y1 = series$TS_2, emDim = p2$optimDim, emLag = p2$optimLag, emRad = NA, targetValue = 0.05)
rn_plot(RN2)
# Get RQA measures
rqa2 <- rp_measures(RN2, silent = FALSE)
# Create RN graph
g2 <- igraph::graph_from_adjacency_matrix(RN2, mode="undirected", diag = FALSE)
V(g2)$size <- degree(g2)
g2r <- make_spiral_graph(g2, arcs = arcs, epochColours = getColours(arcs), markTimeBy = TRUE)
# Get RN measures
rn2 <- rn_measures(g2, silent = FALSE)
# Should be the same
rqa2$RR==rn2$graph_measures$EdgeDensityTS 3
#----------------------
# Adjacency matrix TS_3
#----------------------
plot(ts(series$TS_3))
# Because these are generated signals, look for a drop in FNN below 1%.
p3 <- est_parameters(y = series$TS_3, nnThres = 1)
RN3 <- rn(y1 = series$TS_3, emDim = p3$optimDim, emLag = p3$optimLag, emRad = NA, targetValue = 0.05)
rn_plot(RN3)
# Get RQA measures
rqa3 <- rp_measures(RN3, silent = FALSE)
# Create RN graph
g3 <- igraph::graph_from_adjacency_matrix(RN3, mode="undirected", diag = FALSE)
V(g3)$size <- degree(g3)
g3r <- make_spiral_graph(g3,arcs = arcs ,epochColours = getColours(arcs), markTimeBy = TRUE)
# Get RN measures
rn3 <- rn_measures(g3, silent = FALSE)
# Should be the same
rqa3$RR==rn3$graph_measures$EdgeDensity